
As Ontario continues its gradual easing of COVID-19 public health measures, continued vigilance is necessary for governments to manage the virus in the long-term. A team including members of U of T Engineering’s BioZone is helping to improve alternative monitoring strategies.
One important and cost-effective tool that can help track trends in community transmission, and detect new variants of concern, is wastewater surveillance.
When fecal matter and urine are discharged into a toilet, the bodily fluids can carry viruses, parasites and bacteria, which are then combined in the sewage system before they flow to a wastewater treatment plant.
“This is not a new method. Wastewater surveillance has been used in the past for early detection of other diseases,” says Dr. Minqing (Ivy) Yang (ChemE PhD 1T5), a research associate in the BioZone lab and a member of its SARS-CoV-2 wastewater surveillance team.
“One huge advantage of wastewater monitoring is everyone must use a toilet, so we are able to capture data for both symptomatic and asymptomatic infections,” Yang adds. “Our results don’t depend on an individual’s actions — whether a sick person is going to do a clinical test or not.”
Since December 31, provincial regulations around clinical PCR testing have meant that daily case counts provide an unclear assessment of how widespread the virus is in the population. Wastewater surveillance provides crucial supplementary data: a pooled community sample that can track whether community transmission is increasing or decreasing.
“When we look at the viral signal results, we don’t look at a particular day’s data in terms of ‘low’ or ‘high’ levels. We are interested in the trend, whether the numbers are going up or down,” says Yang. “Overall, we are monitoring wastewater data from 2.7 million people in the Greater Toronto Area, which is about 18% of Ontario’s population.”
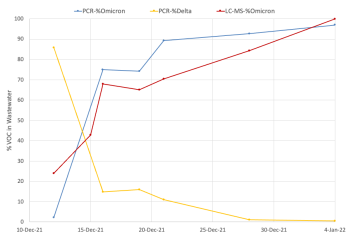
“We were able to successfully track the Alpha wave last spring, as well as the Delta wave late last summer, and the recent Omicron wave in late December to early January,” Yang adds.
Still, this type of critical wastewater testing has a lot of uncharted ground for researchers, says Professor Hui Peng (Chemistry), who co-leads the surveillance team with Professor Elizabeth Edwards (ChemE).
“There has been a lot of progress in this field in the past two years,” he says. “And we expect to continue our studies for at least another year, since we have the funding support from the Ontario Ministry of Environment, Conservation and Parks (MECP).”
While the U of T team monitors wastewater samples from nine treatment plants — three out of the four plants that cover the City of Toronto, and six in the Halton region — they are also working to track new variants of concern and improve the detection of the SARS-CoV-2 viral signals that are often weakened by dilution and environmental factors.
The BioZone lab receives 24-hour composite wastewater samples from the nine sites three times a week. Once it’s concentrated by a technician, RNA is extracted from the sample. Research associates will then conduct a one-step reverse transcription qPCR analysis on the extracts to quantify levels at which the SARS-CoV-2 virus is present.
The technique is the same as a clinical PCR test, only the sample — solids extracted from wastewater — is different.
Within a 48-hour window, the team uploads its data to the MECP’s data hub, where the lab’s partners, Toronto Public Health and Halton Public Health, can access it to inform their public health decisions. Since January 2022, the public health units have made this data publicly available.
There are challenges with this type of environmental sampling that the team is working to resolve. The viral signal in wastewater can be affected by interfering factors, such as inclement weather and the bodily fluids of healthy individuals, which can lower the viral signal and make it less detectable.
Advancing the early detection of variants of concern is also a top priority for the lab.
“Each of the variants of concern have sequences that are very similar, and some mutations are only single-base mutations, which means the RNA sequence is very similar,” says Peng.
In response, Peng and Edwards have developed a mass spectrometry method, which the team believes is the first to be used in the SARS-CoV-2 wastewater surveillance field.
“This method is very powerful in helping us discriminate the variants, even if their sequences are very similar,” says Peng. “We can even use mass spectrometry to discriminate the difference when there is only one single-base mutation on the sequence.”
“We are also trying to improve the sensitivity of the viral signal through the removal of interferences that affect PCR detection. We have not completed that investigation yet, but we have made some promising progress.”
